
Genomics technologies, such as ATAC-seq, ChIP-seq, and DNase-seq, have revolutionised molecular biology, generating a complete genome’s worth of signal in a single assay. The challengeis no longer data generation, it's effectively and reproducibly extracting biological meaning from such massively complex datasets. While other tools approach this problem with simple statistical tests, our novel machine learning model uses a convolutional neural network, local genomic enrichments measurements, and Poisson-based significance testing from multiple viewpoints, all integrated using a multilayer perceptron to give a probability of being a true biological signal. We hand-labelled 499Mb of genomic data, built 5,000 models, and tested with over 100 unique users from labs around the world. And because it’s built on the powerful MLV visualisation software, results can easily be visualised and shared with collaborators or reviewers. The culmination of these efforts is a peak caller that can extract more from your data, create interactive charts, and improve interpretability - all while simplifying the analysis process.
Create a Project

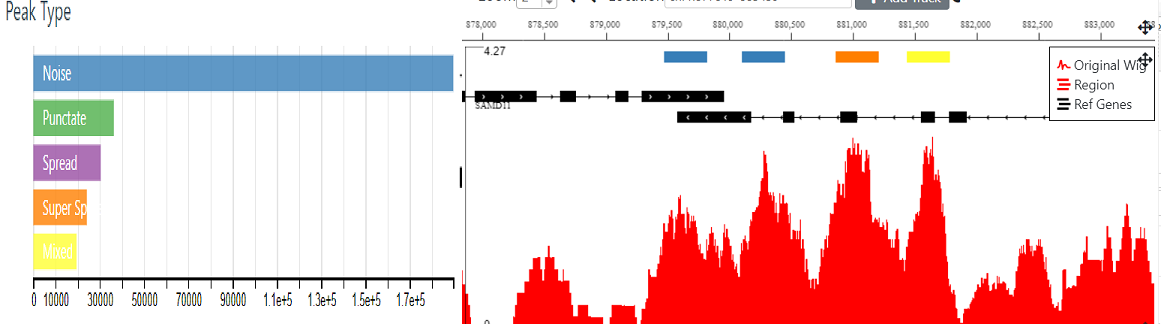
All LanceOtron modules require a bigwig file to supply the model with coverage data.
We recommend directly converting BAM files to bigwigs with deepTools] using the following command:
bamCoverage --bam filename.bam.sorted -o filename.bw --extendReads -bs 1 --normalizeUsing RPKM
The options used in this command are important, as they affect the shape of peaks and therefore the neural network's assessment. Extending the reads out to the fragment length represents a more accurate picture of coverage (N.B for paired end sequencing the extension length is automatically determined, single end tracks will require the user to specify the `--extendReads` length), as does using a bin size of 1 (the `--bs` flag). We recommend RPKM normalisation, as this was also used for the training data.
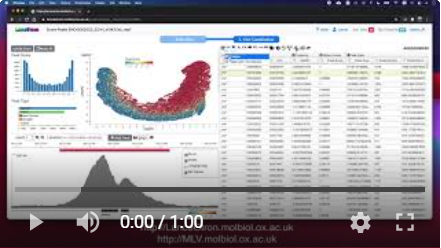
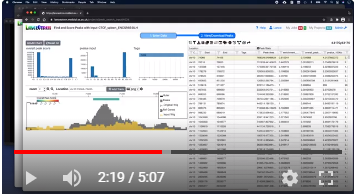
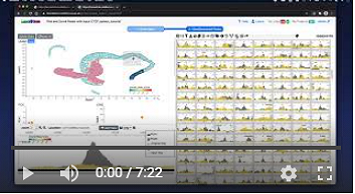
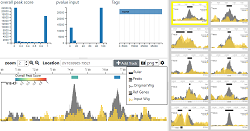
Watch a video demonstrating the basic functionality of LanceOtron